Note
Click here to download the full example code
Time-evolving violin plot¶
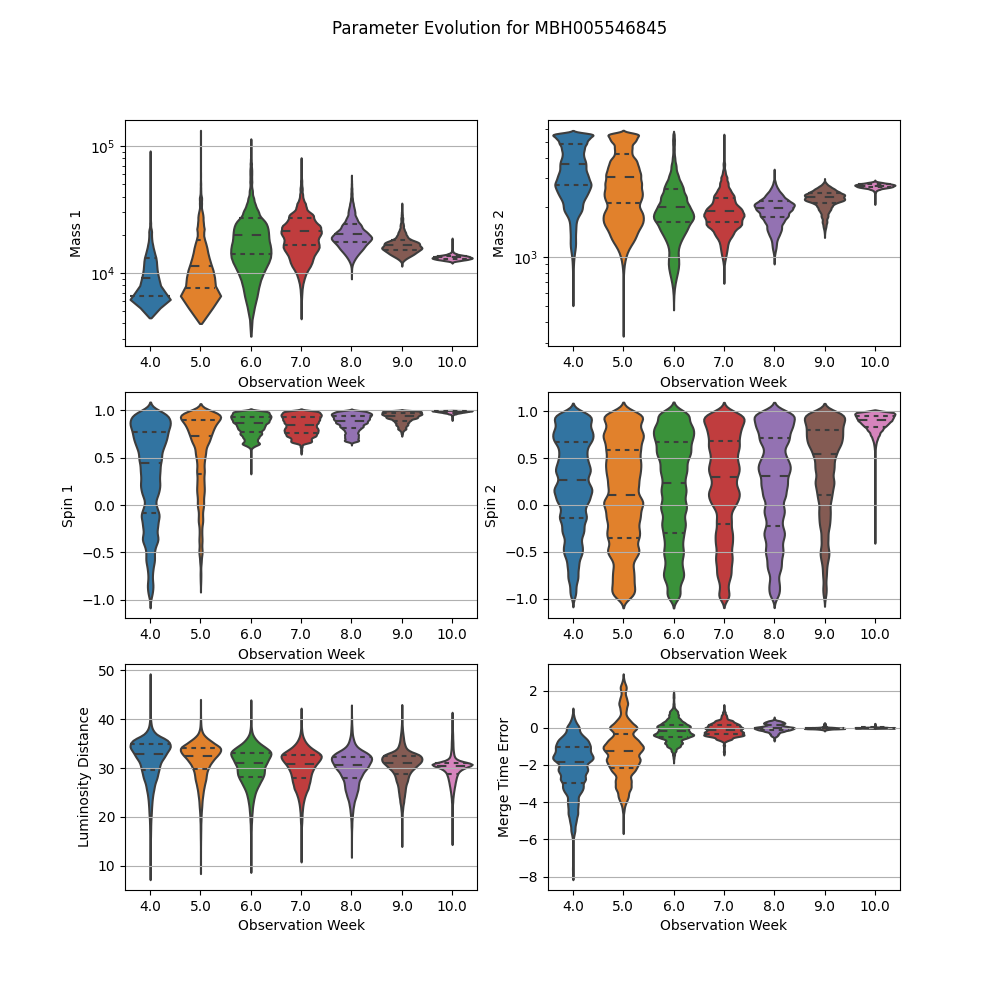
Violin plot of select parameters for a single source showing how parameter estimation changes with observing time over all observing epochs
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from lisacattools.catalog import GWCatalogs
from lisacattools.catalog import GWCatalogType
# get list catalog files
catPath = "../../tutorial/data/mbh"
catalogs = GWCatalogs.create(GWCatalogType.MBH, catPath, "*.h5")
last_cat = catalogs.get_last_catalog()
detections_attr = last_cat.get_attr_detections()
detections = last_cat.get_detections(detections_attr)
detections = detections.sort_values(by="Mass 1")
detections[
["Parent", "Log Likelihood", "Mass 1", "Mass 2", "Luminosity Distance"]
]
Choose a source from the list of detections and get its history through the different catalogs
# Pick a source, any source
sourceIdx = "MBH005546845"
# Get source history and display table with parameters and observing weeks containing source
srcHist = catalogs.get_lineage(last_cat.name, sourceIdx)
srcHist.drop_duplicates(subset="Log Likelihood", keep="last", inplace=True)
srcHist.sort_values(by="Observation Week", ascending=True, inplace=True)
srcHist[
[
"Observation Week",
"Parent",
"Log Likelihood",
"Mass 1",
"Mass 2",
"Luminosity Distance",
]
]
allEpochs = catalogs.get_lineage_data(srcHist)
# For plotting purposes, we add a new parameter that is merger time error (in hours), expressed relative to median merger time after observation is complete
latestWeek = np.max(allEpochs["Observation Week"])
allEpochs["Merge Time Error"] = (
allEpochs["Barycenter Merge Time"]
- np.median(
allEpochs[allEpochs["Observation Week"] == latestWeek][
"Barycenter Merge Time"
]
)
) / 3600
Create the violin plot select the parameters to plot and scaling (linear or log) for each
params = [
"Mass 1",
"Mass 2",
"Spin 1",
"Spin 2",
"Luminosity Distance",
"Merge Time Error",
]
scales = ["log", "log", "linear", "linear", "linear", "linear"]
# arrange the plots into a grid of subplots
ncols = 2
nrows = int(np.ceil(len(params) / ncols))
fig = plt.figure(figsize=[10, 10], dpi=100)
# plot the violin plot for each parameter
for idx, param in enumerate(params):
ax = fig.add_subplot(nrows, ncols, idx + 1)
sns.violinplot(
ax=ax,
x="Observation Week",
y=param,
data=allEpochs,
scale="width",
width=0.8,
inner="quartile",
)
ax.set_yscale(scales[idx])
ax.grid(axis="y")
# add an overall title
fig.suptitle("Parameter Evolution for %s" % sourceIdx)
plt.show()
Total running time of the script: ( 0 minutes 16.736 seconds)